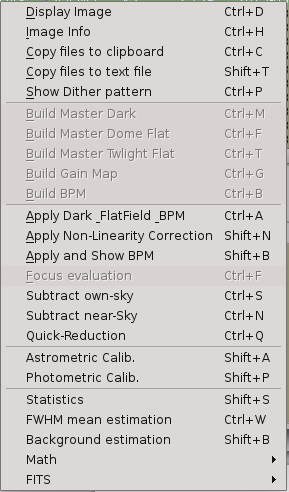
2. PANIC Quick-Look Tool (PQL)¶
2.1. Purpose¶
PANIC Quick-Look (hereafter PQL) performs some on-line data processing for quick-look or quality check of the data being acquired, taking a close look at a raw near-infrared image and getting a quick feedback of the running observation.
PQL is an application with a graphical user interface which monitors the GEIRS data output, waiting for new FITS files coming from GEIRS. When a new file is detected, it is added to the file list view in the main panel, and then PQL will perform the task previously specified by the user in the setup configuration. Some of the available tasks are:
Only display the FITS image with no processing
Dark subtraction, flat division
Sky subtraction (using N-nearest frames or own sky )
Field distortion removal
Image align and stacking
Preliminary astrometric solution
Preliminary photometry
In addition, PQL allows you to execute manually in an interactive way some tasks with the data. For example, you will be able to select a file, compute some statistics values (background, FWHM, min, max, …) or ask for the sky subtraction looking for the nearest N frames around the selected one. Other option available is to select a set of files and request to shift and align them.
PQL can be operated in both near-real time mode (during the observation) and offline mode (after the observation, with all data files already stored in the disk); however, its functionalities have been provided mainly in near-real time to check the status and progress of the observation during the night.
The visualization application used to display the images is SAOImage ds9, which supports FITS images, multiple frame buffers, region manipulation, and many scale algorithms and colormaps.
2.2. FITS files and headers¶
PQL only supports FITS (Flexible Image Transport System) with two-dimensional
image formats. Because PANIC has a FPA of four detector, the FITS files can be Single Extension FITS (SEF)
or Multi-Extension FITS (MEF), however MEF are prefered.
The complete definition of the FITS headers can be found on the GEIRS documentation.
For general purpose, such as viewing and simple analysis, only minimal headers keywords are required. However, and in order to group and reduce observing sequences, the following header keywords are also required:
OBS_TOOL= 'OT_V1.1 ' / PANIC Observing Tool Software version
PROG_ID = ' ' / PANIC Observing Program ID
OB_ID = '6 ' / PANIC Observing Block ID
OB_NAME = 'OB CU Cnc Ks 2' / PANIC Observing Block Name
OB_PAT = '5-point ' / PANIC Observing Block Pattern Type
PAT_NAME= 'OS Ks 2 ' / PANIC Observing Secuence Pattern Name
PAT_EXPN= 1 / PANIC Pattern exposition number
PAT_NEXP= 5 / PANIC Pattern total number of expositions
IMAGETYP= 'SCIENCE ' / PANIC Image type
These keywords are automatically added to the FITS header by the PANIC Observation Tool, as each file is created. If these are not saved, PQL will not work correctly.
2.3. Starting PQL¶
To start PQL GUI, you can lauch it from the PANIC computer (panic22/panic35) once you are logged as obs22. Thus, as any one of the workstations of the observing room, open a X terminal window and log into the PANIC computer as follow:
for 2.2m:
$ ssh -X obs22@panic22
(ask Calar Alto staff for password)
Once you are logged into the PANIC computer, to launch PQL GUI type next command:
$ start_ql &
The next figure shows a snapshot of the main window of PQL GUI that will bring up the start_ql command.
2.4. Configuration files¶
The configuration files used by PQL are located in the $PAPI_HOME/config_files. The main config file is the same file used by PAPI, ie., $PAPI_CONFIG, and usually called papi.cfg.
This file includes a lot of parameters used by PAPI, and therefore by PQL during the processing; however at the end of the $PAPI_CONFIG file there is section called quicklook, where the user can set some specific parameters for PQL:
##############################################################################
[quicklook]
##############################################################################
# Next are some configurable options for the PANIC Quick Look tool
#
# some important directories
#
source = /data1/PANIC/
output_dir = /data2/out # the directory to which the resulting images will be saved.
temp_dir = /data2/tmp # the directory to which temporal results will be saved
verbose = True
# Run parameters
run_mode = Lazy # default (initial) run mode of the QL; it can be (None, Lazy, Prereduce)
Although the user can edit these values in the config file, some of them can be set easily on PQL’s GUI.
For the complete list of the parameters available on the $PAPI_CONFIG file, see Main config file section.
2.5. PQL’s main window¶
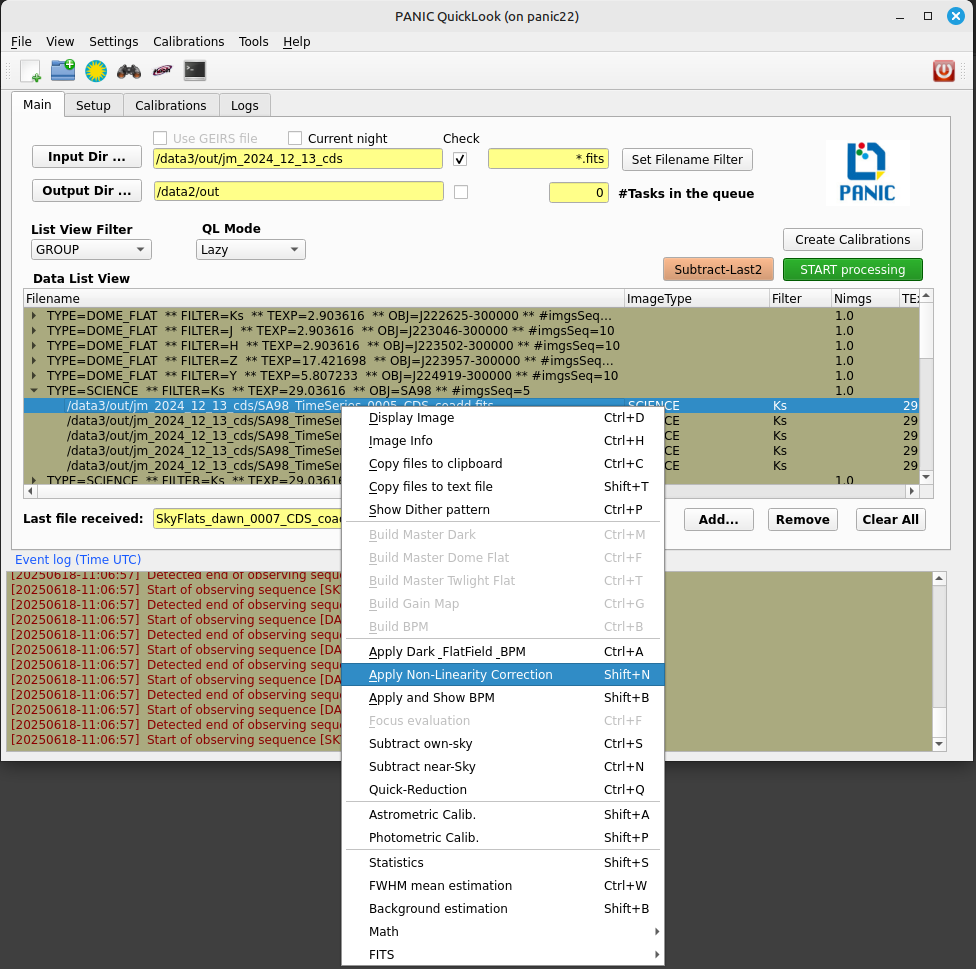
PQL Main window contains a Menu bar (1), Tool bar (2), four Tabbed panels (3) and an Event Log Window (4). Images are displayed in an external well-known application, ds9. Plots results are displayed in the additional windows, usually generated by matplotlib than can be copied to the clipboard, printed or saved.
2.5.2. Tool bar¶
The tool bar duplicates some of the options available from the menu bar or the pop-up menu. Currently, there are several buttons which provide quick access to change the most frecuently-used PQL actions:
add a file to the current view
change the source input directory: the same that Input directory.
display the current selected image: the same that Display.
open an IRAF console
open Aladin tool
quit PQL (on the right border)

2.5.3. Main panel¶
This tab panel contains the following controls:
Input directory
Ouput directory
Filename filter
Current night
Use GEIRS file
Data list view
List view filter
QL mode
‘Subract last-2’ button
‘START processing’ button
‘Create Calibrations’ button
2.5.3.1. Data directories¶
In the ‘Main’ tab panel of PQL main window, the fitst thing to set up are the data directories:
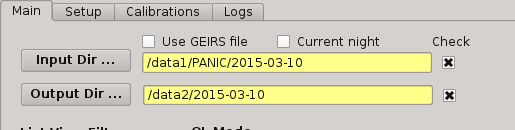
2.5.3.2. Input directory¶
This is where you tell PQL where the data are or being saved by GEIRS. This directory is specified at the beggining of the night on the Observation Tool. PQL requieres all data to lie in some main directory, not being required to distribute the files in individual sub-directories for darks, flats, and science images. It is advised that this directory follow the next format:
/data1/PANIC/YYYYMMDD
To set the value, the user must push the ‘Input Dir’ button:

Note that the value in this field has only effect when the checkbox on the right is clicked.
2.5.3.3. Output directory¶
This is where you tell PQL where the data generated by PQL, as result of some processing, will be saved. This directory must also be specified at the begining of the night, and is advised to follow the next format:
/data2/out/YYYYMMDD
To set the value, the user must push the ‘Output Dir’ button:

Note that the value in this field has only effect when the checkbox on the right is clicked.
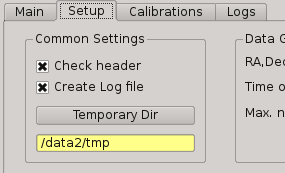
2.5.3.4. Temporal directory¶
This is where you tell PQL where the temporal files generated by PQL, as result of some processing, will be saved, and probably deleted after at the end of that processing. This directory must also be specified at the begining of the night, and is advised to follow the next format:
/data2/tmp/YYYYMMDD
To set the value, the user must push the ‘Temporary Dir’ button than appears on the ‘Setup’ tab, instead the ‘Main’ tab used for input and output directory.
2.5.4. Current night checkbox¶
When you click this checkbox, the Input directory and Output directory fields will be automatically filled with the currect night date. If the current night Input/Ouput directories donot exist, PQL will ask you if you want to create them.
The currect night is supposed to start at 8 am (UTC) and to end at 8 am (UTC) of next day.
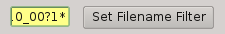
2.5.5. Filename filter¶
In this box, the user can filter the name of the files should appears on the data list view from the input directory (output files are not filtered). The filter can contains ‘*’ and ‘?’ wildcards.
For example:
*March10_00?1*
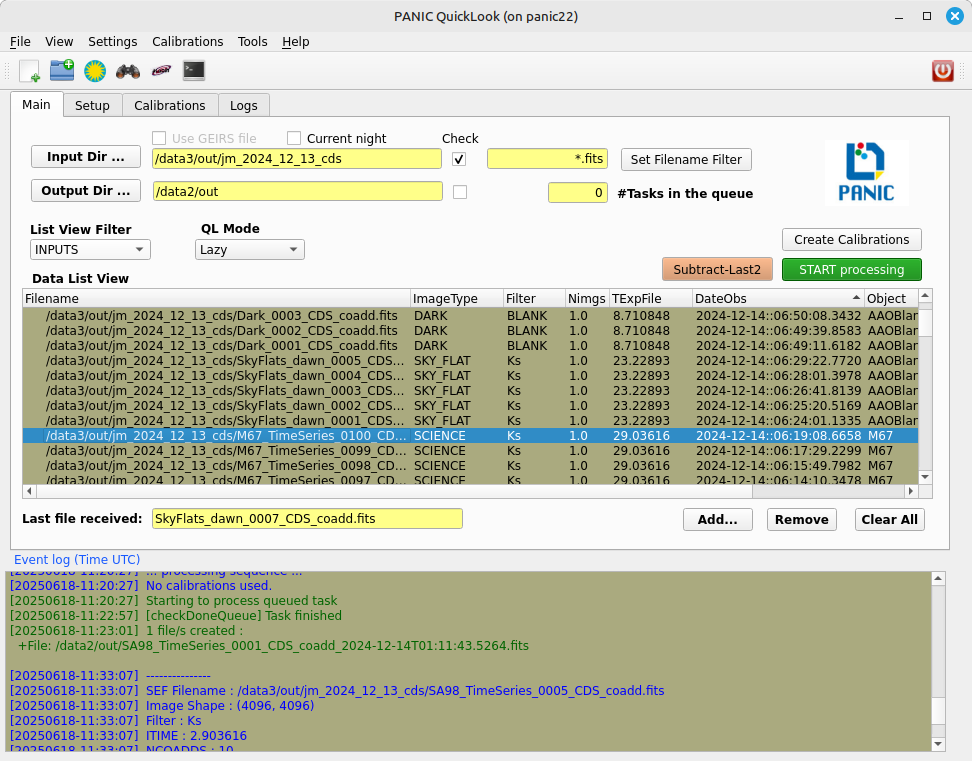
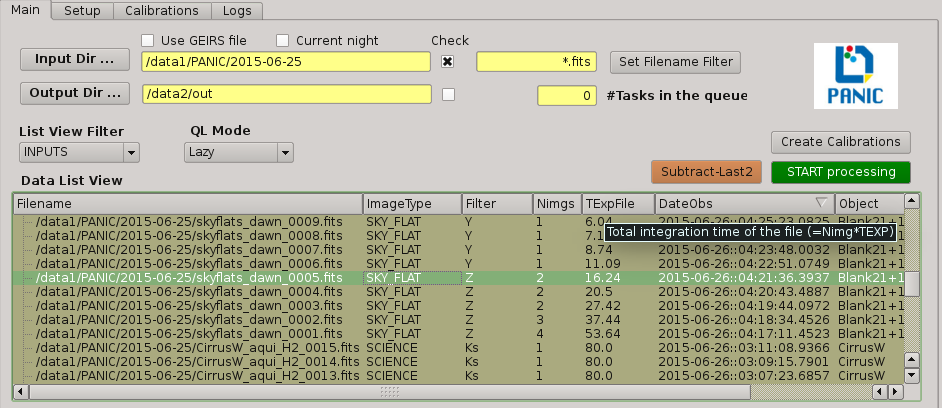
2.5.6. Data list view¶
Tha data list view control displays all the files found in the input directory, or in the output directory if the check box at the right of output directory is checked. Additionaly, the use can add any other FITS file. The control is a multicolum table with the next fields:
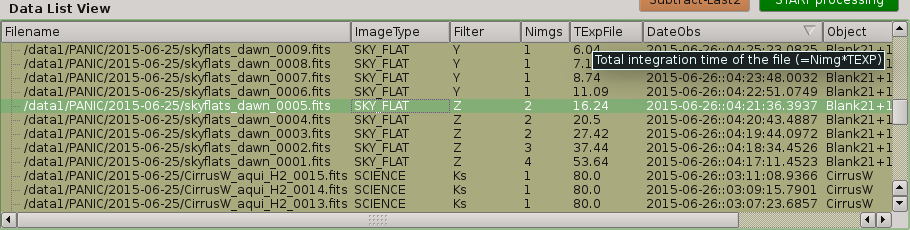
- Filename
Full path name of the file found in the
- Image type
The type of the FITS file detected: DARK, DOME_FLAT, SKY_FLAT, FOCUS, SCIENCE
- Nimgs
Number of images (layers) of the cube; if image is integrated (no cube), then = 1.
- TExpFile
Total Exposition time of the file (= Nimgs * EXPTIME) (Thus, EXPTIME = TExpFile / Nimgs)
- Date-Obs
Observation data of the file (DATE-OBS keyword)
- Object
Object name (OBJECT keyword)
- RA
Right ascention of center of the image.
- Dec
Declination of the cener of the image.
You can sort the list by any column (filename, image type, exptime, filter, date-obs, object, right ascension, declination) by clicking on their headers, as usual; by default, the list is sorted by the Date-Obs field, showing the most recect file at the top.
A double-click on any row displays all its file into SAOImage ds9.
For further details of any of the files, you can also look at the header of a fits image using ds9 using the “File/Display Fits Header…” menu option.
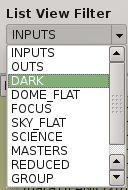
2.5.7. List view filter¶
It allows you to select the type of files to be shown in the data list view. The options are:
- INPUTS
Files of the input directory
- OUTS
Files of the ouput directory
- DARK
Files marked (IMAGETYP) as DARK images
- DOME_FLAT
Files marked as DOME_FLAT image
- FOUCS
Files marked as FOCUS image from a focus series
- SKY_FLAT
Files marked as SKY_FLAT images
- SCIENCE
Files marked as SCIENCE image or with unknown type.
- MASTERS
Files marked as MASTER calibration files produced by PAPI
- REDUCED
Files marked as calibrated by PAPI
- GROUP
Special case that show all the files groupped as observed sequences (OBs)
- ALL
Show all the files, not matter the type of it
2.5.8. QuickLook mode¶
The quick look mode combo box allows you to select the mode in which PQL will be run when the START processing button is pushed. The current modes are:
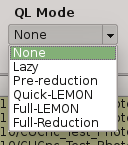
- None
No processing action is done
- Lazy (default)
If the end of a calibration (DARK, FLAT) sequence is detected, the master file is built. Otherwise, the SCIENCE files are processed as specified in the ‘Setup->Lazy Mode’:
Apply DARK + FLAT + BPM
Subtract Last Frame (Science)
Subract Nearest Sky
- Pre-Reduction
If the end of observing sequence is detected, it is processed in a quick mode (single pass for sky subtraction). For calibration sequences, the master file will be built, and for science sequences, a quick reduction will be done using options configured in the ‘Setup->Pre-Reduction Mode’ and the calibrations found in local database (output directory and external calibration directory). Note that the pre-reduction options configured in the config file will be overwritten.
- Quick-LEMON
The same as Pre-reduction, but the processing stops after the 1st sky subtraction, and no final co-added image is produced. It is useful for LEMON processing for light curves.
- Full-Reduction
If the end of observing sequence is detected, it is processed in a science mode (double pass for sky subtraction). For calibration sequences, the master file will be built, and for science sequences, a science reduction will be done using options configured in the ‘Setup->Pre-Reduction Mode’ and the calibrations found in local database (output directory and external calibration directory). Note that the pre-reduction options configured in the config file will be overwritten.
- Full-LEMON
The same as Pre-reduction, but the processing stops after the 2nd sky subtraction, and no final co-added image is produced. It is useful for LEMON processing for light curves.
2.5.9. Last file received¶
This field shows the last file received (detected) by PQL.
2.5.11. Setup Panel¶
This panel allows the user to set some of the parameters used for the processing. It is divided into six group boxes as shown in next figure:
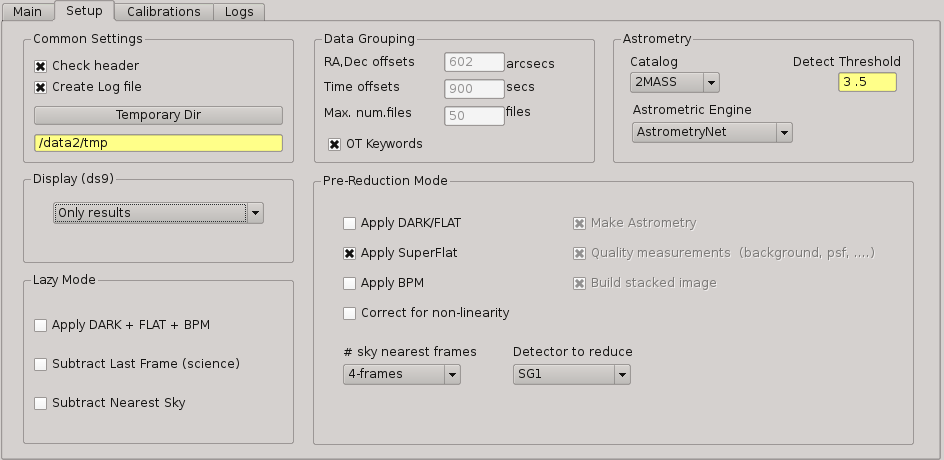
2.5.11.1. Common Settings¶
In this group you can set the next parameters:
Check header
Create log file
Temporary directory
2.5.11.2. Data grouping¶
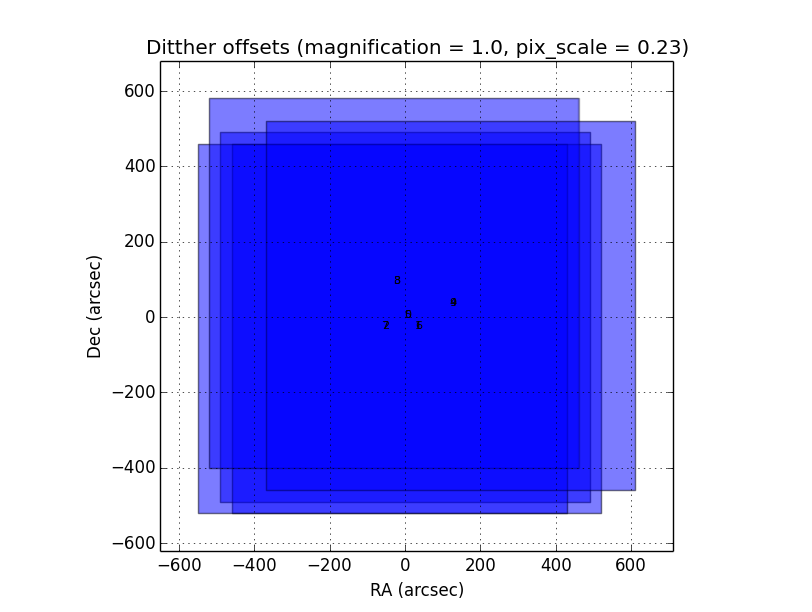
It contains some parameters used for the data grouping when any OT keywords are present; in that case, PQL will try to group the files follwing the near in sky and time criterion:
RA,Dec offsets:
Time offsets:
Max. number of files:
If OT keywords are present, then check box ‘OT’ should be ckecked (default mode).
2.5.11.3. Astrometry¶
In this group you can set some parameters related with the astrometric calibration done during the processing:
Catalog: reference catalog used for the calibration (2MASS , USNO-B1, GSC 2.2, SDSS-R5, GAIA (up to DR3))
#. Astrometric Engine: which tool you want to use to the astrometric calibration (SCAMP or Astrometry.net). For the case of Astrometry.net, you can use 5200 index files (GAIA-DR2), which are the default ones, or you can use 4200 index files (2MASS).
Detect threshold: the SExtractor threshold to be used to detect sources
2.5.11.4. Display¶
Here you can select which files are displayed automatically in the DS9. You have next options:
Only results (default): only FITS files created in the output directory as result of some processing
Only new files: only new FITS files detected in the input directory
All files: both new files detected in the input directory and the results in the output directry.
None: no files will be displayed
2.5.11.5. Lazy mode¶
Under this box, the user can select the operations to be executed when the Lazy Mode is activated in PQL. Currectly, the available and exclusive operations are:
Apply Dark + Flat + BPM
Subtract Last Frame (science)
Subtract Nearest Sky
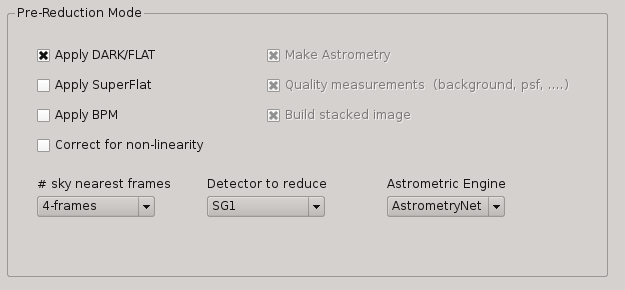
2.5.11.6. Pre-reduction¶
Under this box, the user can select the operations to be executed when the Pre-reduction Mode is activated in PQL. Currectly, the available and exclusive operations are:
Apply Dark and FlatField
Apply SuperFlat (default)
Apply BPM (Bad Pixel Map)
Correct for non-linearity
Select the number of frames to computer the sky bacground: 1-5 (default 4)
Detector to reduce: SG1 (default), SG2, SG3, SG4, SG123, All
2.5.12. Calibrations panel¶
This panel allows the user to set some of values for the search of master calibration files.
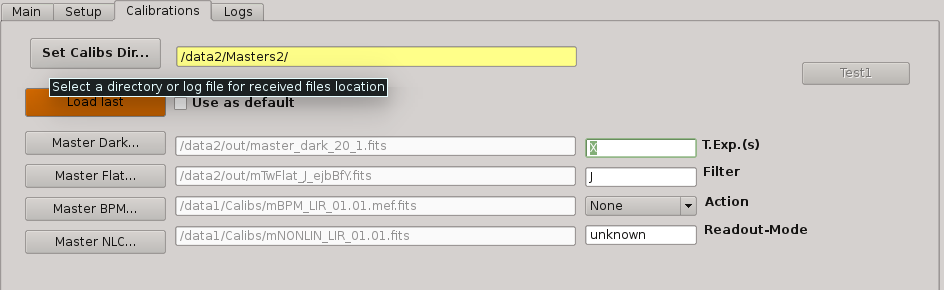
2.5.12.1. Set Calibs Dir¶
Pushing this button the user select the additional (external) directory from which the QL will look for master calibration files. Normally, it is used to provide to the QL with additional calibrations (dark, flat) from previous nights. Master calibrations found in the output directory will have higher priority than those ones.
This directory is also called ‘external calibration’ in PAPI command line:
-C EXT_CALIBRATION_DB, --ext_calibration_db=EXT_CALIBRATION_DB
External calibration directory (library of Dark & Flat
calibrations)
Or ext_calibration_db in the config file.
Then, if during the reduction of a ReductionSet(RS) no calibrations (dark, flat) are found in the current RS, then PAPI will look for them into this directory. If the directory does not exists, or no calibration are found, then no calibrations will be used for the data reduction. Note that the calibrations into the current RS have always higher priority than the ones in the external calibration directory.
2.5.12.2. Load last¶
When this button is pushed, the most recent master calibration files found in output directory and external calibrations are shown in the fields below.
If Use as default is click-checked, then the displayed files will be used as default calibrations when Apply Dark_FlatField_BPM is run. Otherwise, Apply Dark_FlatField_BPM routine will ask the user for the master calibration files to be used.
2.5.13. Log panel¶
It is an extension or duplicate of the Even Log window of the main panel, but with a wider area for messages.
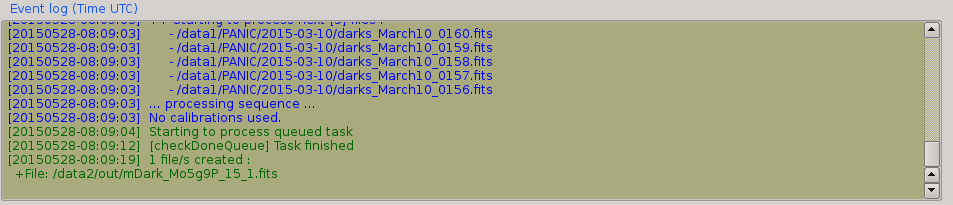
2.6. Event log window¶
The event log window shows important events and results generated by PQL. For example, the filename of the new files generated are shown, or the error produced while the processing of some sequence. This window is used only as output, and you cannot type any command on it.
2.8. How do I …?¶
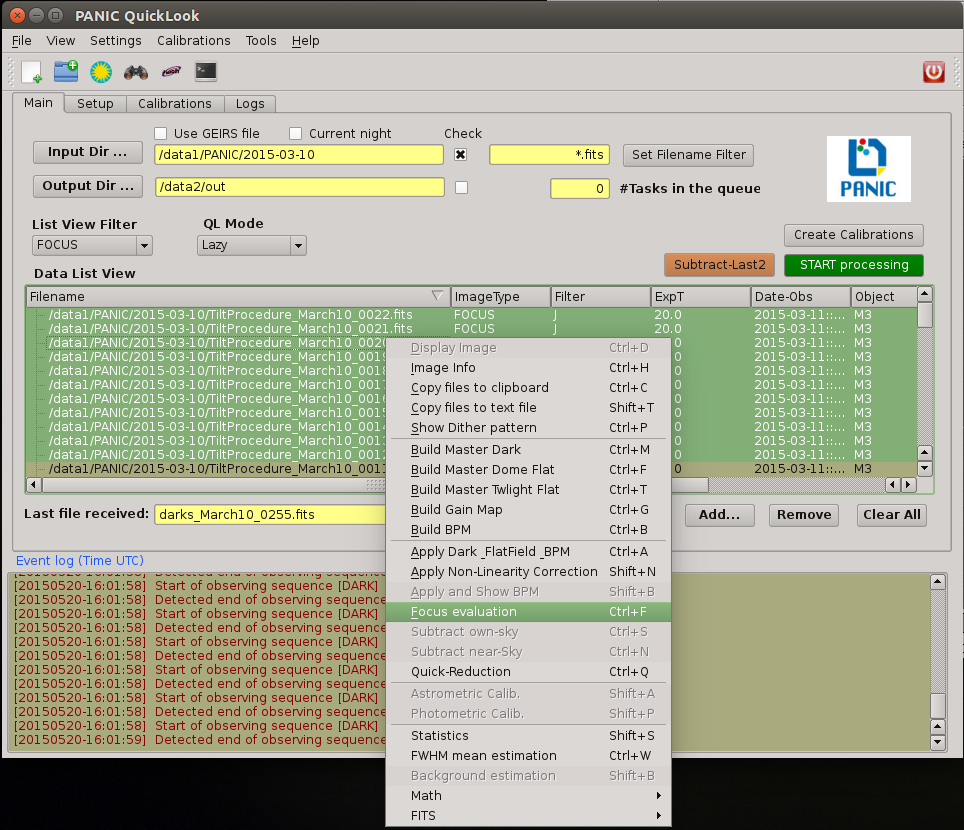
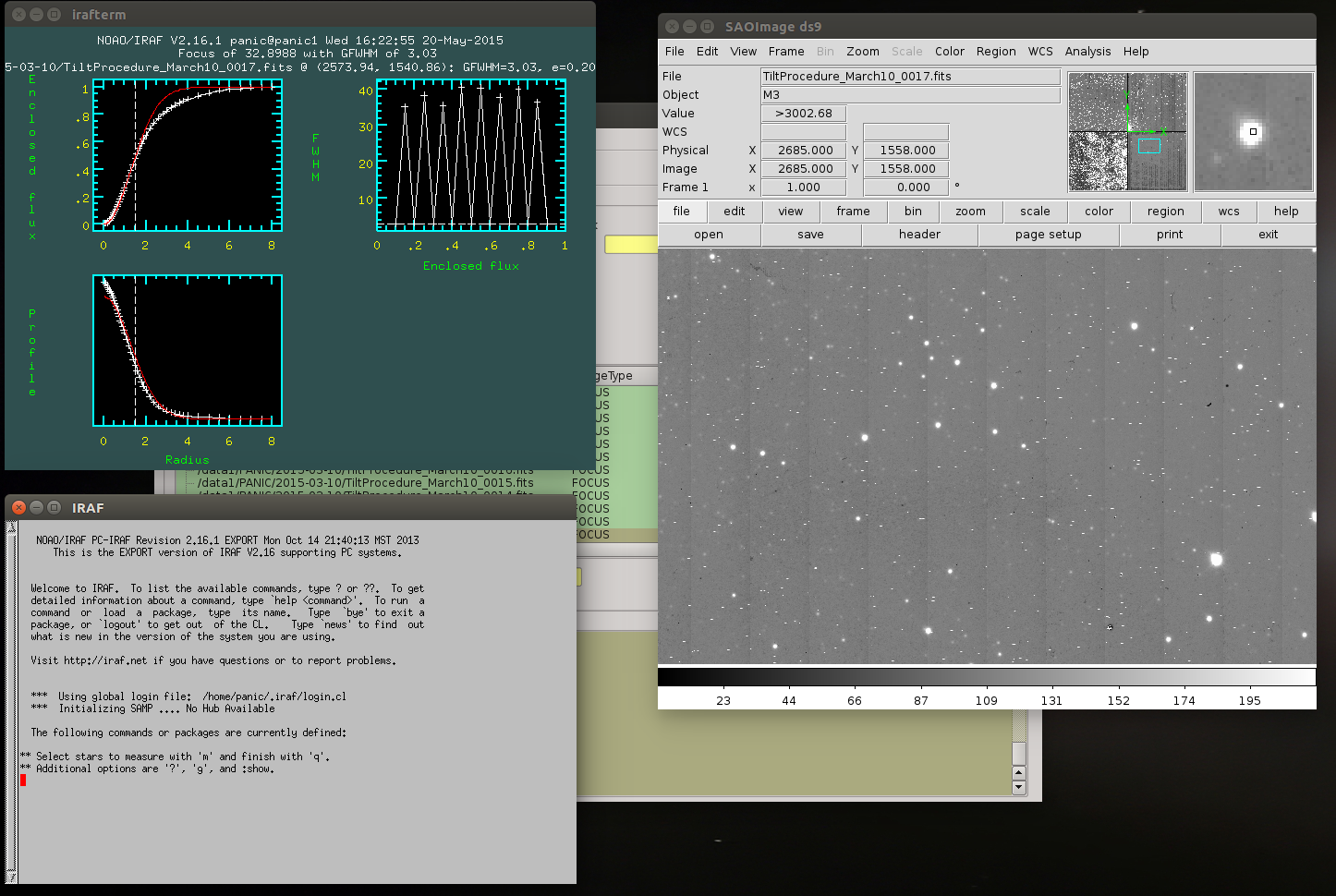
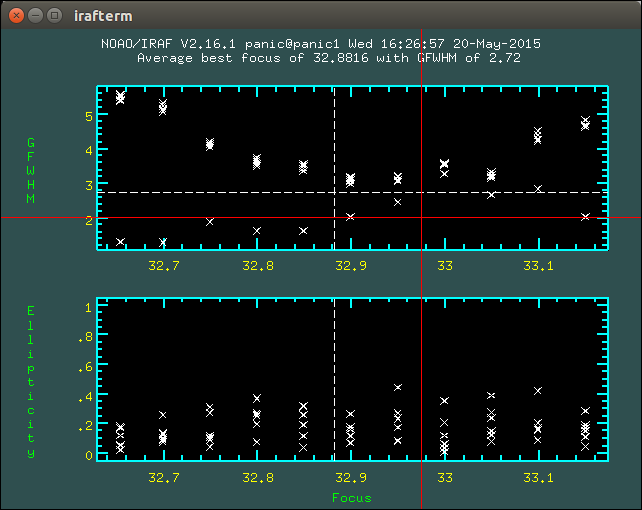
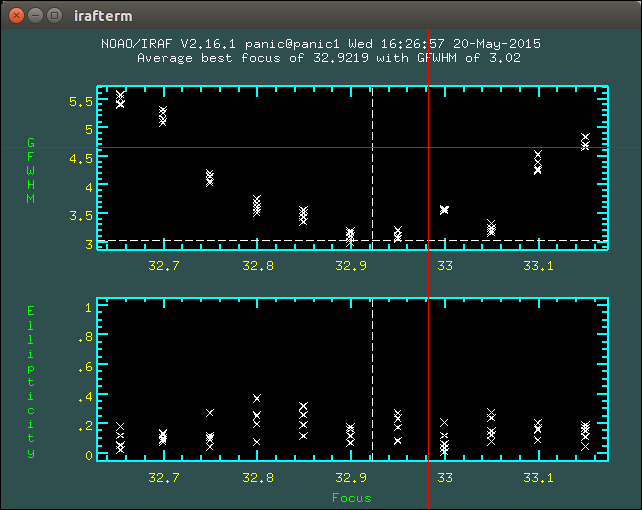
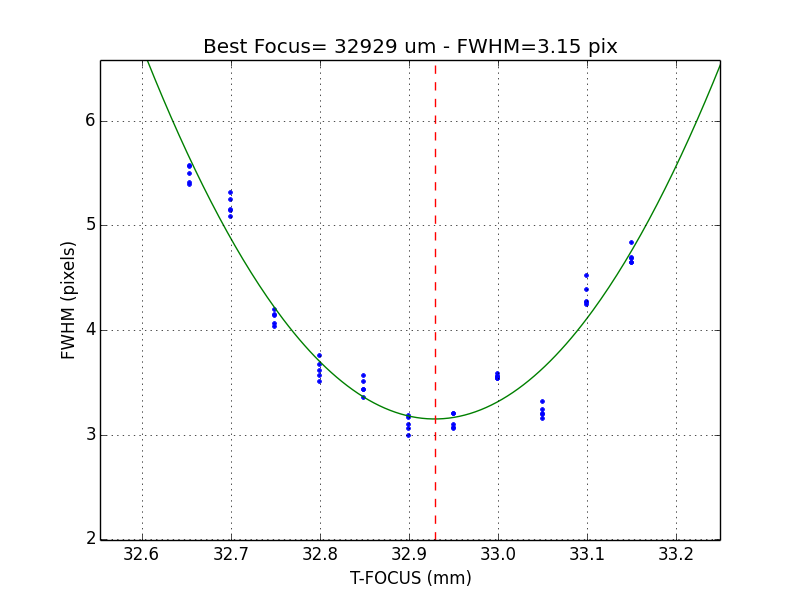
2.8.1. How to determine the telescope focus ?¶
To determine the telescope focus, you should run a OT focus serie around the guest value and then run the Focus Evaluation.
2.8.2. How to determine the field rotation ?¶
To determine the field rotation, firstly you should observe a enough crowded field and then run the astrometric calibration on it for each detector. Once you have the new FITS astrometrically calibrated, you have to look for the ROTANGLE keyword in the new header. For example:
ROTANGLE= -0.032836 / degrees E of N
2.8.3. How to inspect the profile of the stars in an image ?¶
You should follow the next steps:
select in the Data List View the image to inspect.
double-click to display the image into ds9 and zoom to the area you wish to inspect
go to the tool bar (or Tool menu) and open an IRAF console
type in the iraf console ‘imexam’
focus the mouse cursor on the ds9 display and type the imexam comand you wish for the inspection. For example, type *r* to show the radial profile of the selected star
once you have finished the inspection, type q to exit from imexam
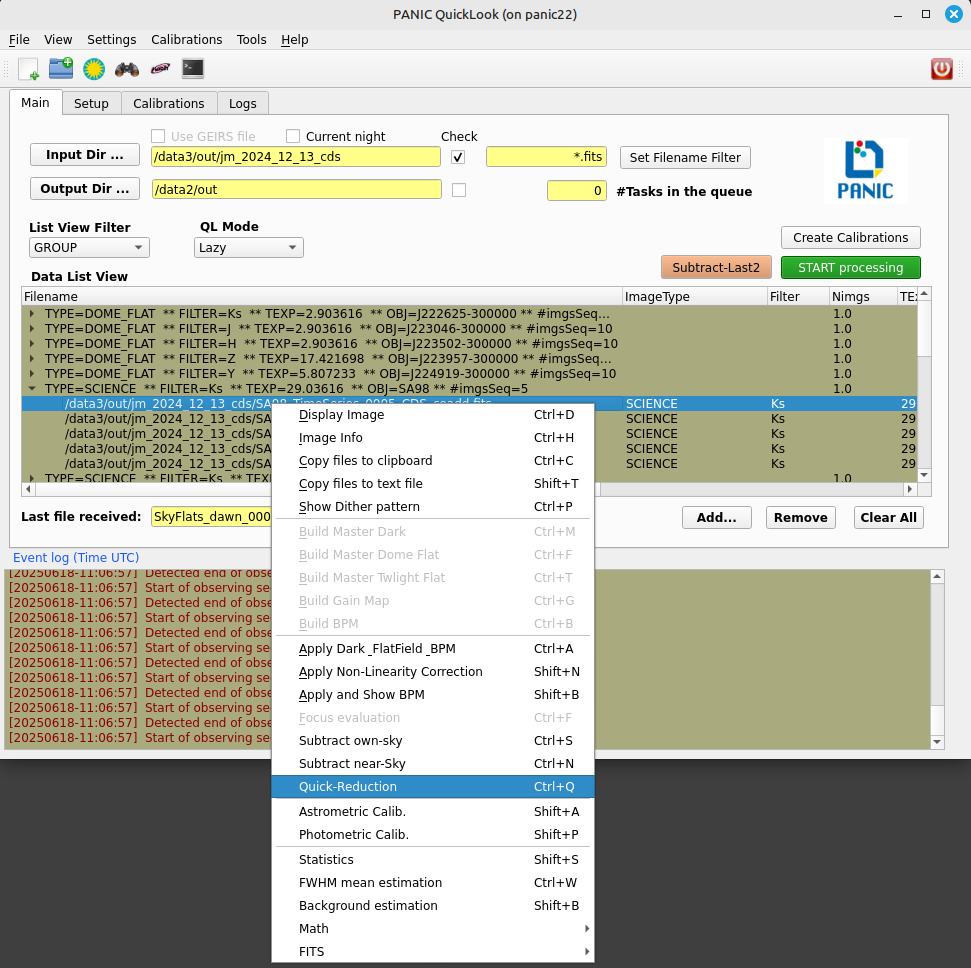
2.8.4. How do I quick-reduce an observed sequence ?¶
There are two options:
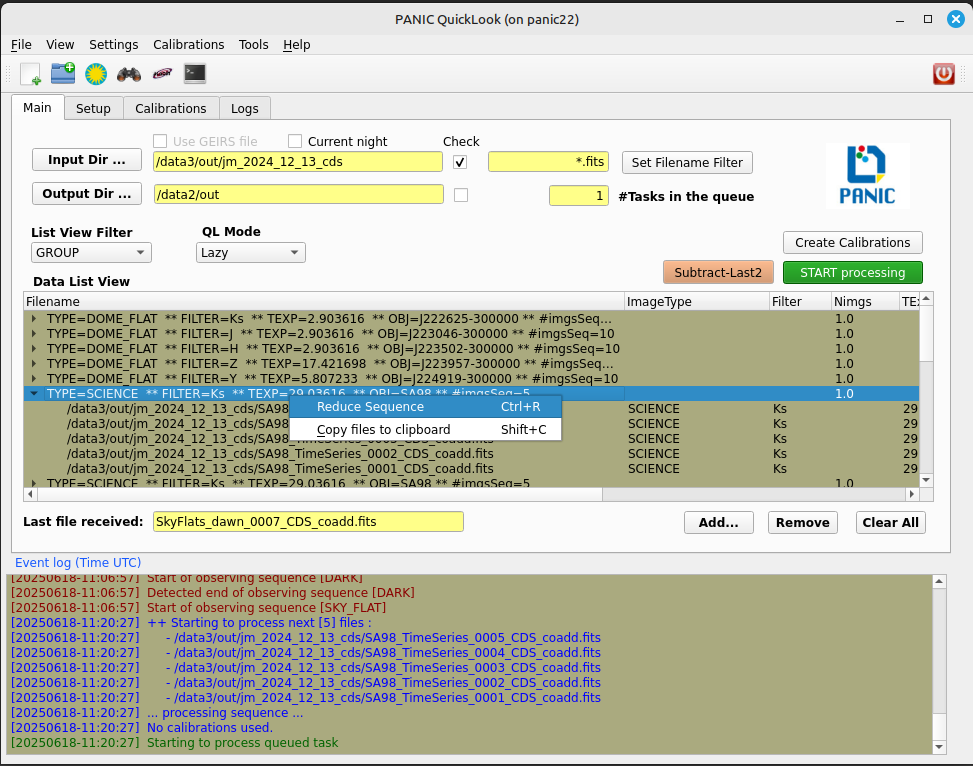
if you know the files that compose the sequence, you can select them and then right-click and run the Quick-Reduction command.
go to the List View Filter and select GROUP; then look for the sequence you are looking for in the Data List View, right-click and select Reduce Sequece command.
For the quick reducion, the pipeline will use the preferences established on ‘Setup’ tab.
2.8.5. How do I quick-reduce an observed sequence using dark and flat master calibration files ?¶
You should follow the next steps:
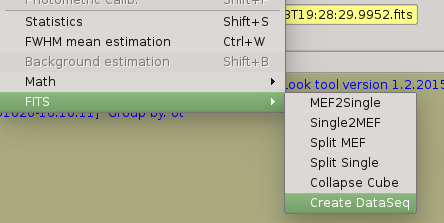
1. Check your sequences are right,ie., they are well-formed and there were no interruption. It some sequece (calibration or science) is not well-formed, the you should use ‘FITS->Create DataSeq’ menu option in order to fix not well-formed sequence.
Create the output directory for the calibrations; then create the calibration pushing ‘Create calibrations’ button in the main panel.
3. When ‘Create calibrations’ have finished, go to ‘Calibration’ tab and select the directory having the master calibrations created just in setep #2.
4. Go to the ‘Setup->Pre-reduction Mode’ tab and check the option ‘Dark/Flat’ and select the detectors you want to process (SG1-SG4).
5. Finally, select the sequence you want to reduce, either selecting one by one the files in the Data List View or selecting the sequence with the ‘Group’ classification; then run ‘Quick-reduction’ from the Pop-up menu.
2.8.6. How do I make mosaics with PQL?¶
By default, PQL proccess or pre-reduce only the SG1 detector (Q1), and then no mosaic is built. However, you can go to the Setup Tab and modify in the Detector to reduce combo box the detector/s to reduce; in case of selecting All or SG123 (all less SG4), the corresponding mosaic will be generated.
Currently, PAPI aligns and coadds (using SWARP or Montage, see mosaic_engine in config file) the images as they are located on the sky to build the mosaic.
2.8.7. How do I make use of parallelisation ?¶
Just be sure the number of parallel parameter is set to True on the $PAPI_CONFIG file. When parallel=True, the pipeline will reduce each detector in parallel using all the cores available in your computer. It is only available for the case of having data from the older PANIC H2RGdetectors (SG1-SG4).
2.8.8. How do I report a issue ?¶
Please submit issues with the issue tracker on github.