9. Troubleshooting¶
This section gives a description of some problems that can be obtained and how they can be solved.
9.1. Why do I get a “command not found” error when trying to run one of the PAPI modules?¶
If you are trying to run the command from the same directory as the executable (i.e.,$HOME/bin), make sure that your path contains “.”
If you are trying to run the command from another directory, add $HOME/bin to your path.
9.2. PAPI does not recognize a group of files as a well formed sequence.¶
It maybe because the sequence is missing some files of the sequence, or some of them have no proper headers (ie. they were not observed with OT).
Solutions:
You can try to reduce the sequence using the groping mode ‘filter’ instead of ‘ot’. It means PAPI will not use any OT header information, but RA, DEC, DATE-OBS and FILTER.
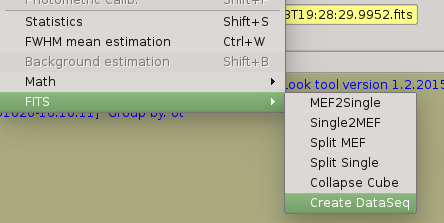
Use the ‘Create Data Seq’ option which will create/fix a new sequence adding the required header keywords:
9.3. PAPI can not run the astrometric calibration¶
Verify that:
You have cdsclient installed
You have an internet connection and no firewall is blocking CDSClient
You can config CDSClient to run with a proxy.
9.4. PAPI can not run the photometric calibration¶
Verify that:
You have an Internet connection. It is used to query the on-line 2MASS or USNO-B catalog.
Check that the input reduced image is astrometrically calibrated and it the filter name FILTER keyword match some of the 2MASS or USNO-B catalog.
9.5. PAPI says some files in the input file list does not look a FITS file.¶
Verify that:
The input file is not ending with a blank/empy line.
The file has Unix text format. Text files created on DOS/Windows machines have different line endings than files created on Unix/Linux. DOS uses carriage return and line feed (”rn”) as a line ending, which Unix uses just line feed (”n”).
9.6. What is the best way to reduce PAPI data?¶
The recommend to use the OT and execute the OBs. That way the headers will include meta-data about the observation, and thus the pipeline can group the data and find the required calibrations for a successful reduction. Then, you only have to type:
> papi -s /data1/PANIC/my_program/
9.7. How can we reach hundredth of magnitude accuracy in photometry ?¶
The best way to accurately photometrically calibrate PANIC images is to use 2MASS stars in the field itself to derive the photometric solution. The accuracy strongly depends on the number of bright 2MASS stars within the filed of view, but ranges from a few 1/100th of a magnitude to 0.1 magnitudes if only faint stars are contained in the field. Additionally, observing supplementary standard star fields can be asked for when preparing the observations. To perform the 2MASS photometric calibration on an image you should use the ‘photometry’ command as follow:
> photometry -i /directory/prereducedField.fits -o test.pdf
9.8. How good is PAPI astrometry and how are PSF variations corrected ?¶
At present the pipeline applies a correction for PSF distortions based on a distortion map derived during the astrometric calibration done with SCAMP (a software developped by Emmanuel Bertin) and 2MASS.
9.9. Is there a way to look at or edit FITS headers?¶
The PAPI package includes the WCS library and tools, including a program called edhead. This is built automatically when you build PAPI, and it is installed into the $HOME/bin. directory.
To run edhead, simply type edhead filename.fits. It will strip the header from the FITS file and open it for editing using the program defined in your environment. Make the changes, save and exit the editor, and the header is re-attached to the image.
9.10. Does PAPI generate a log file of the processing ?¶
Yes, it can be configured in the $PAPI_CONFIG file with the parameter logfile = /tmp/papi.log. For each, execution, the log filename will have an suffix with the timestamp of the data and time, i.e., /tmp/papi_YYYY-MM-DDTHH:MM:SS.ss.log.